Training models for dense prediction tasks, such as segmentation, require a large amount of detailed annotation. This burden is even heavier in histopathology, where tissues within the same class can vary widely in appearance, boundaries are often ambiguous, and even experts frequently disagree.
Active learning (AL) helps by letting a model decide which samples should be labeled next. Instead of labeling everything, you focus expert time on the most informative parts of the dataset. With the right strategy, AL can reach full-annotation performance using only a fraction of the labels.
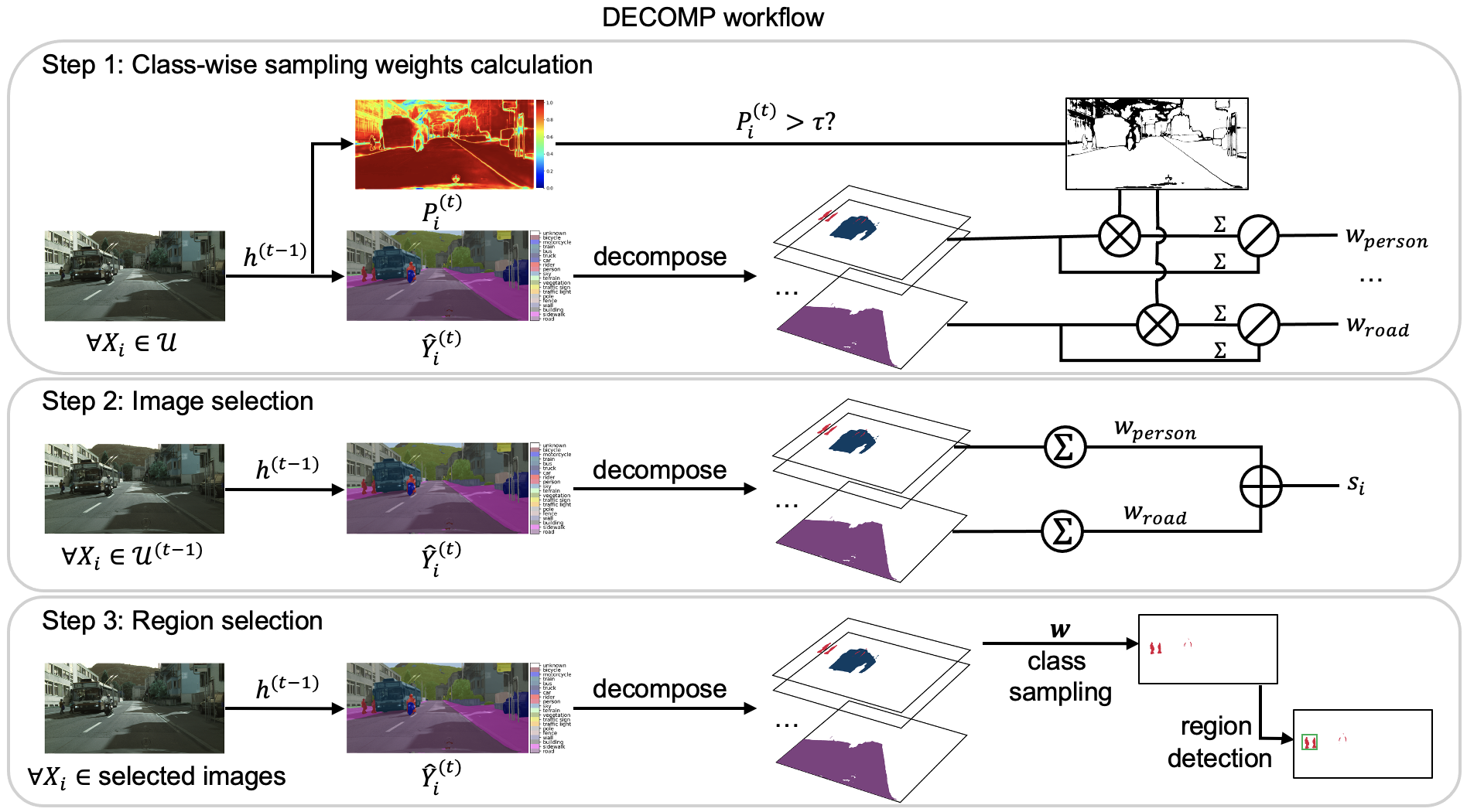
Prior work has demonstrated that labeling regions is more effective than labeling the entire image when applying AL to segmentation tasks. Existing diversity-based methods select representative regions by computing features for each region, then clustering them or computing a core-set that maximally covers the feature space. However, when applied to large medical images (e.g., 3-D CT scans or gigapixel WSIs), these methods run into several problems:
- High computational cost: Every region in every image needs feature extraction and storage.
- Irrelevant regions: Feature diversity does not guarantee task relevance.
- No awareness of model weaknesses: All regions are treated equally, even when the model struggles with specific classes.
To mitigate this, pipelines often add an uncertainty-filtering step, selecting the most uncertain regions before applying diversity sampling. That reduces the computational load but harms diversity because uncertain regions can be similar.
A New Approach: Decomposition Sampling (DECOMP)
We introduce DECOMP, a new AL strategy to select diverse annotation regions, it avoids all the limitations above while being extremely easy to implement.
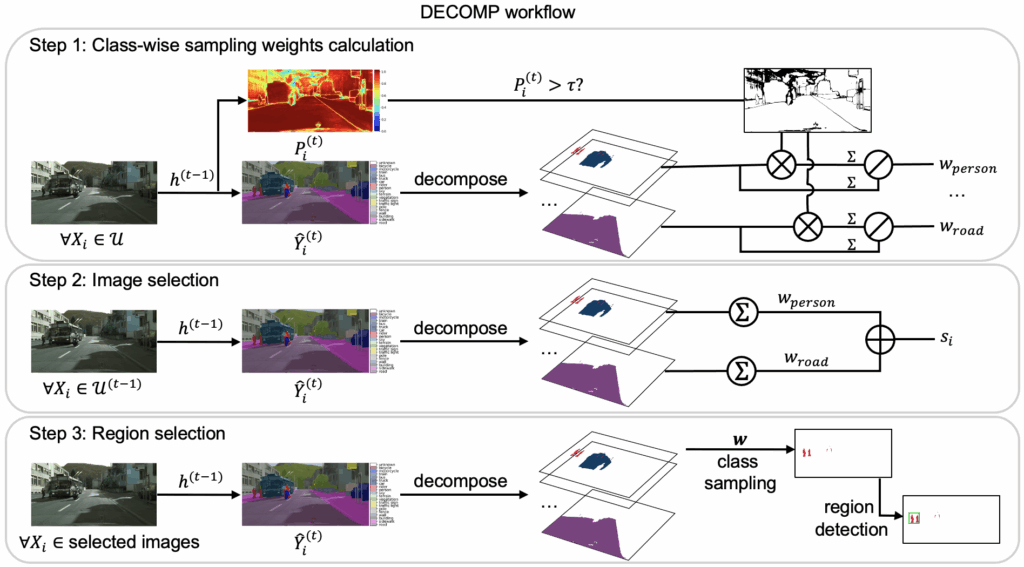
How DECOMP works
- The model predicts the class of every pixel/voxel in an image.
- The image is decomposed into class-specific components using these predictions.
- DECOMP estimates class-wise confidence:
- Classes the model is uncertain about get a higher sampling weight.
- Images containing more high-weight classes receive higher priority.
- For each selected image, DECOMP samples regions class by class, ensuring:
- broad class coverage
- additional annotations for difficult classes
All of this requires only one forward pass per image. No feature banks. No recomputation for overlapping patch features. No clustering. And DECOMP naturally supports regions of arbitrary shape and scales to more classes without additional overhead.
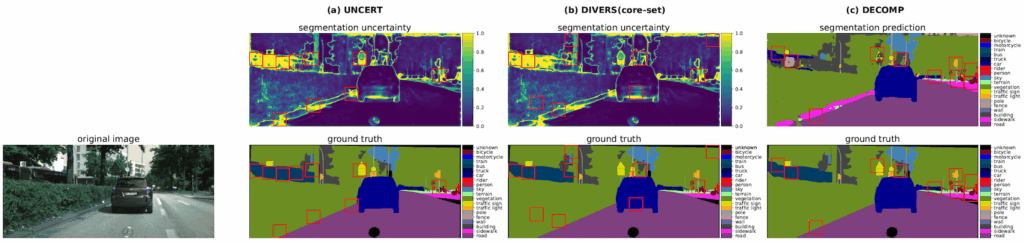
What We Evaluated
We tested DECOMP across three different tasks and observed substantial annotation savings:
- Histopathology ROI classification (BRACS):
Achieves 95% of full-annotation performance while labeling <40% of whole-slide images, with at most 15 ROIs per slide. - 2-D semantic segmentation (Cityscapes):
Achieves 95% of full-annotation performance with annotations covering only 2.5% of the total area. - 3-D kidney/tumor segmentation (KiTS23):
Achieves 95% of full-annotation performance with annotations covering just 0.15% of the total volume.
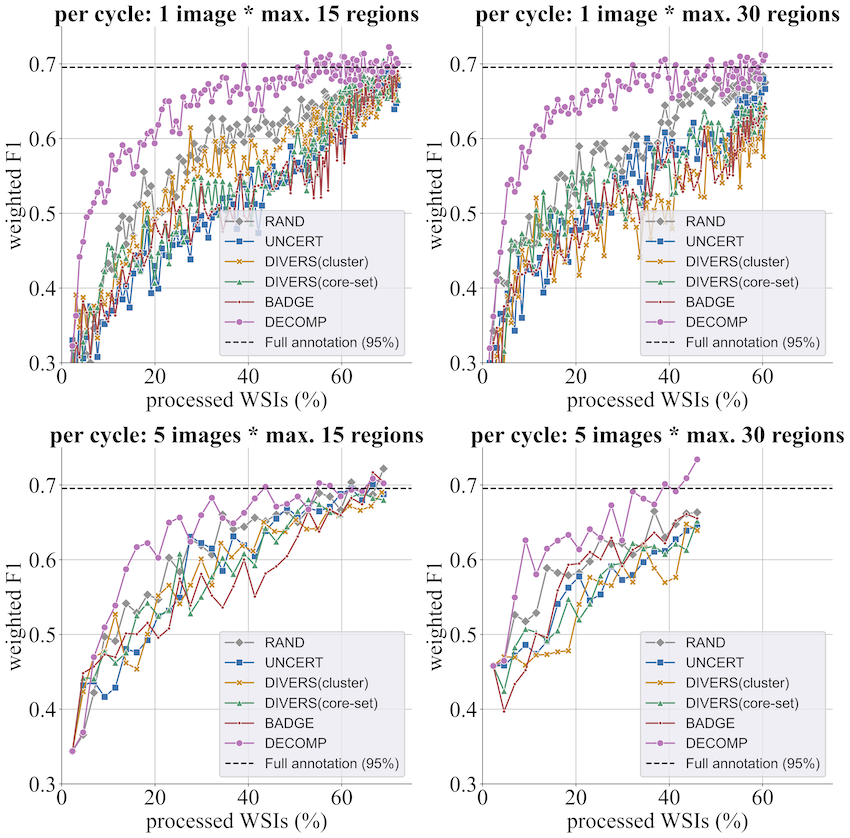
These results highlight DECOMP’s potential for drastically reducing annotation cost in both medical and non-medical settings.
Check out our full paper (published at WACV 2026) for more results, ablation studies, and detailed analysis!
Acknowledgement: d.hip campus – Bavarian aim, DFG project 460333672 CRC1540 EBM, project 405969122 FOR2886 Pandora, projects 505539112, 520330054 and 545049923, and HPC resources from NHR@FAU.


Comments are closed