In the rapidly evolving field of computational pathology, foundation models (FMs) have become the new hot topic. These models, trained on millions of unlabeled tissue tiels using self-supervised learning, promise a “universal” understanding of histopathology. But how do they perform on high-precision, fine-grained tasks like mitotic figure classification?
In our recent study published in the Journal of Machine Learning for Biomedical Imaging (MELBA), titled Benchmarking Foundation Models for Mitotic Figure Classification, we conducted a systematic evaluation of these models. We did not just want to know which model is “best” but rather we wanted to understand how they scale with data and how they handle the “domain shift” that often plagues clinical AI adoption, all in comparison to standard supervised training of common vision models.
The Experiment: 6 Pathology FMs, 2 Adaption Strategies, 2 Massive Datasets
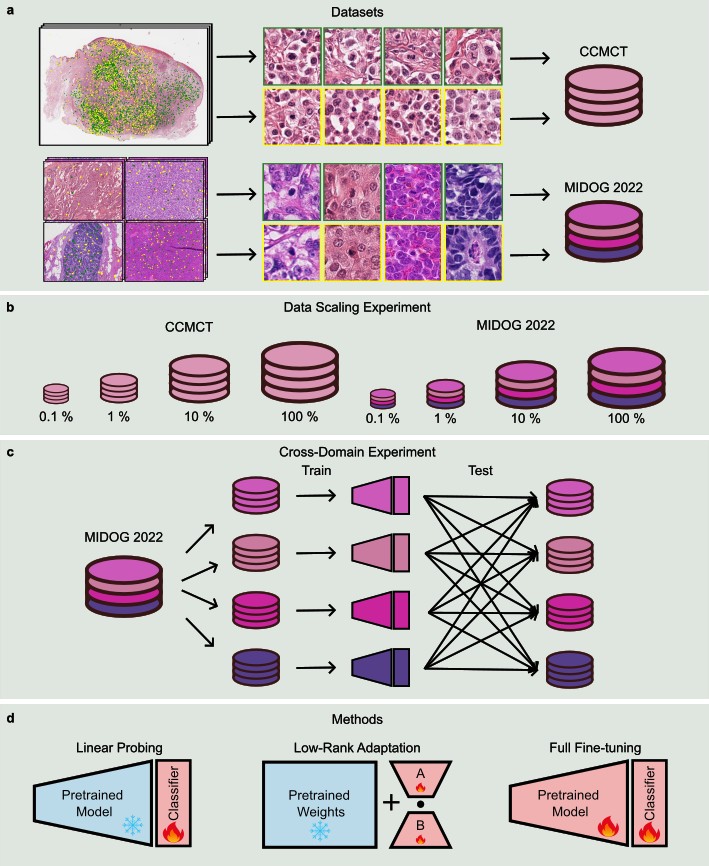
We benchmarked six state-of-the-art pathology foundation models: Phikon, UNI, Virchow, Virchow 2, H-optimus-0, and Prov-GigaPath. These models represent a spectrum of architectures (from ViT-B to ViT-G) and training scales (from 43 million to over 2 billion tiles).
To test their limits, we utilized two distinct datasets:
- CCMCT: A large-scale, single-domain dataset of canine cutaneous mast cell tumors with over 40k mitotic figure annotations.
- MIDOG 2022: A highly diverse, multi-tumor, multi-species dataset designed to test generalization across different laboratories and scanners.
We compared three primary training paradigms:
- Linear probing: Training only a classification head while keeping the foundation model frozen.
- Low-Rank Adaptation (LoRA): A parameter-efficient fine-tuning method that adapts the model’s internal attention mechanisms.
- End-to-End Baselines: Traditional CNNs (ResNet50) and ViTs trained from ImageNet weights.
Result 1: The Efficiency of LoRA
Our data scaling experiments revealed a clear hierarchy. While Linear Probing is computationally “cheap,” it consistently underperforms. In contrast, LoRA-adapted models showed incredible data efficiency.
On the CCMCT dataset, models like H-optimus-0 and Virchow2 achieved AUROC scores near 0.90 with only 10% of the training data. This suggests that the quantity of labels is potentially no longer a bottleneck for pathology labs, but rather the strategy used to adapt models.
Result 2: Conquering the Domain Shift
The “Holy Grail” of medical AI is a model that works as well in a new hospital as it did in the one where it was trained. Our cross-domain experiments on the MIDOG 2022 dataset showed that:
- End-to-End training and linear probing saw strong performance drops when evaluating un unseen tumor types.
- LoRA-adapted FMs (specifically Virchow 2 and H-optimus-0) almost entirely closed the performance gap. They maintained an out-of-domain AUROC of 0.87-0.88, demonstrating strong robustness of combining massive scale-pretraining with efficient fine-tuning techniques.
Result 3: The “Baselines” Still Bite
One surprising takeaway was the resilience of the ResNet50 End-to-End baseline. During the data-scaling experiments and on the in-domain evaluation of the cross-domain experiments, a fully fine-tuned ReNet50 often outperformed foundation models that were only adapted via linear probing. This highlights a critical lessen: Foundation models are not a “magical bullet” unless they are properly adapted. To beat a well trained traditional model, you must allow the foundation model to fine-tune its internal represenations, which is exactly where LoRA excels.
Conclusion: A new Blueprint for Pathology AI
Our findings support the growing consensus that foundation models, when paired with efficient adaptation strategies, are poised to transform computational pathology by enabling robust, scalable solutions that generalize across tasks and domains. Instead of the labor-intensive process of training task-specific models from scratch, the most robust and efficient path might be to:
- Focus on high quality, diverse data.
- Select a high-capacity pathology foundation model.
- Apply Low-Rank Adaptation rather than simple linear probing.
Future research should extend these benchmarks to additional tasks, datasets, and adaptation methods, and explore strategies for further improving out-of-domain generalization.
Read the full paper here: https://doi.org/10.59275/j.melba.2026-a3eb


Comments are closed