Insights from the 2022 edition of the MIDOG MICCAI Challenge
At MICCAI 2022, we held the second edition of the MItosis DOmain Generalization (MIDOG) challenge. The challenge was about detecting mitotic figures (depicting cells undergoing cell division) in histopathology images. These cells are an important marker and play a significant role in the assessment of malignancy of many tumors.
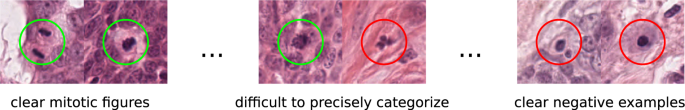
Given the importance of this task, it might surprise at first hand that the inter rater discordance of experts in identifying those cells is rather low. This is, however, also one of the reason why it’s a task that is well worth automating. In the first (2021) edition of the MIDOG challenge, we showed that robust solutions (working on different digitization devices) are viable in the case of one of the most prevalent tumors – breast cancer.
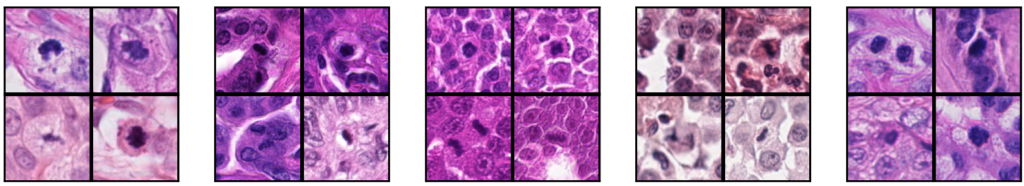
The MIDOG 2022 challenge widened the scope of this task to mitotic figure detection across various tumor types, including 6 types for training and 10 in the independent test set. Visually, these look remarkably different, which also explains the deterioration of most previous models on this task:
Today, the preprint of the challenge report was released. Click on the image to see it, or scroll down below to get a brief summary.
A brief summary of insights
Insight 1: Model optimization is key.
While very different strategies were employed for the task in the MIDOG challenge, also involving partially interesting new ideas to tackle with the problem of domain generalization, the winning team of the challenge (from the TIA Center of the University of Warwick) performed a rather straight-forward two-stage approach, which they optimized really well.
Insight 2: Robust mitosis detection on various domains is possible today.
While in previous years, detection of mitotic figures was a major challenge for automatic methods, especially in adverse real-world conditions, our challenge shows that given the proper amount of (highly variable) training data, deep learning methods can generalize well to new domains. This shows a possible path towards application in clinical practice, which we assume is about to set in soon.
Insight 3: Immunohistochemistry-aided annotation is not necessarily in line with expert consensus on H&E
We established our challenge ground truth, as usually done, using a multi expert consensus. While our challenge confirms again, that this is a viable thing to do, we alternatively also evaluated a ground truth that was created using the H&E stain but also, additionally, a re-stained version of the slide using PHH3 stain. This additional information led to a much more precise definition of mitotic figure, but also revealed a considerable number of previously undistinguishable cellular events that were found to be actual mitotic figures. For a ground truth (in the typical definition of the word), we thus can summarize that it is sensible to go for IHC-aided toolchains.



No responses yet