When Precision Meets Ambiguity:
Information Mismatch in PHH3-Assisted Mitosis Annotation Leads to Interpretation Shifts in H&E Slide Analysis
Histopathology is a very important part of diagnosing and understanding tumors, with mitotic figure (MF) counts serving as a cornerstone in assessing tumor aggressiveness. But what happens when the tools we use to enhance precision unintentionally alter our understanding? In our recent study, we investigated this question, focusing on the interplay between traditional hematoxylin and eosin (H&E) staining and immunohistochemical, mitosis-specific phospho-histone H3 (PHH3) staining.
The Context: Why This Matters
In histopathology, the identification and quantification of mitotic figures (MFs) is a key metric for assessing tumor proliferation and aggressiveness. This count, typically performed on hematoxylin and eosin (H&E)-stained slides, plays a central role in tumor grading systems and ultimately informs clinical decision-making. However, the task is notoriously challenging, as it relies heavily on the pathologist’s ability to discern subtle morphological features indicative of mitosis. Low inter-rater agreement is a well-documented issue, with pathologists frequently disagreeing on which cells should be classified as MFs.
Phospho-histone H3 (PHH3) immunohistochemical staining offers a potential solution to this problem. PHH3 is a specific marker for mitosis, binding selectively to histone proteins during cell division. This specificity makes PHH3 an attractive adjunct to H&E, as it highlights mitotic cells with high contrast, even in cases where morphological cues might be ambiguous. Studies have shown that PHH3-stained slides not only reduce inter-rater variability but also increase the overall recall of mitotic figures, as the stain reveals cells in various stages of mitosis, including early phases that might otherwise be overlooked in H&E. By using PHH3-assisted annotations as a reference, pathologists can cross-validate their H&E-based assessments, potentially generating a more consistent dataset.
This perceived reliability has led to a growing interest in PHH3-assisted annotation workflows, particularly in the context of machine learning (ML) applications. It is assumed that PHH3-generated annotations offer a higher-quality ground truth because they more closely reflect the biological reality of mitosis, independent of the variability introduced by subjective morphological interpretation. By leveraging PHH3 as a guide, pathologists can identify mitotic figures that might have been misclassified or missed entirely in H&E slides, reducing noise in the training datasets used for ML models.
However, while PHH3 has the potential to enhance precision, it also introduces questions: Is the information in both stains the same? And what are the effects of training with a ground truth based only on H&E compared to a ground truth assisted with PHH3?
The Study: A Rigorous Experimental Approach
We designed a series of experiments to explore this question from multiple perspectives [1]. In all experiments, we used images derived from the exact same glass slides, which were initially stained with H&E, then de-stained and re-stained with PHH3. This approach allowed us to obtain corresponding representations of each cell in both stains.
- In a large-scale study, pathologists annotated MFs in H&E-stained sections both with and without the support of PHH3. This setup enabled us to directly measure the impact of PHH3 assistance on the pathologists’ annotations. Furthermore, MFs identified exclusively through PHH3 assistance were re-evaluated by a panel of pathologists to determine whether these MFs could also be reliably identified in H&E staining alone.
- The datasets generated from this study were then used to train and evaluate different detection models, allowing us to assess how ground truths created with and without PHH3 assistance influence model performance. Among these models was a dual-stain detector, which received paired PHH3-stained and H&E-stained patches as input. By providing the detector with the same information available to the pathologists during PHH3-assisted annotation, we aimed to replicate and evaluate its ability to integrate complementary data from both stains.
Overview of the PHH3-assisted annotation procedure. During the annotation process, pathologists could view the corresponding PHH3-stained slides as a transparent overlay on their H&E-stained counterparts.
Key Findings: A Double-Edged Sword
In our results we found that:
- Improved Inter-Rater Agreement: PHH3-assisted annotations significantly boosted consistency among experts, with object-level agreement metrics (F1-score) jumping from 0.53 to 0.74, and inter-rater reliability reached near-perfect levels (ICC = 0.99). This was somewhat expected as it is in line with other works in this field.
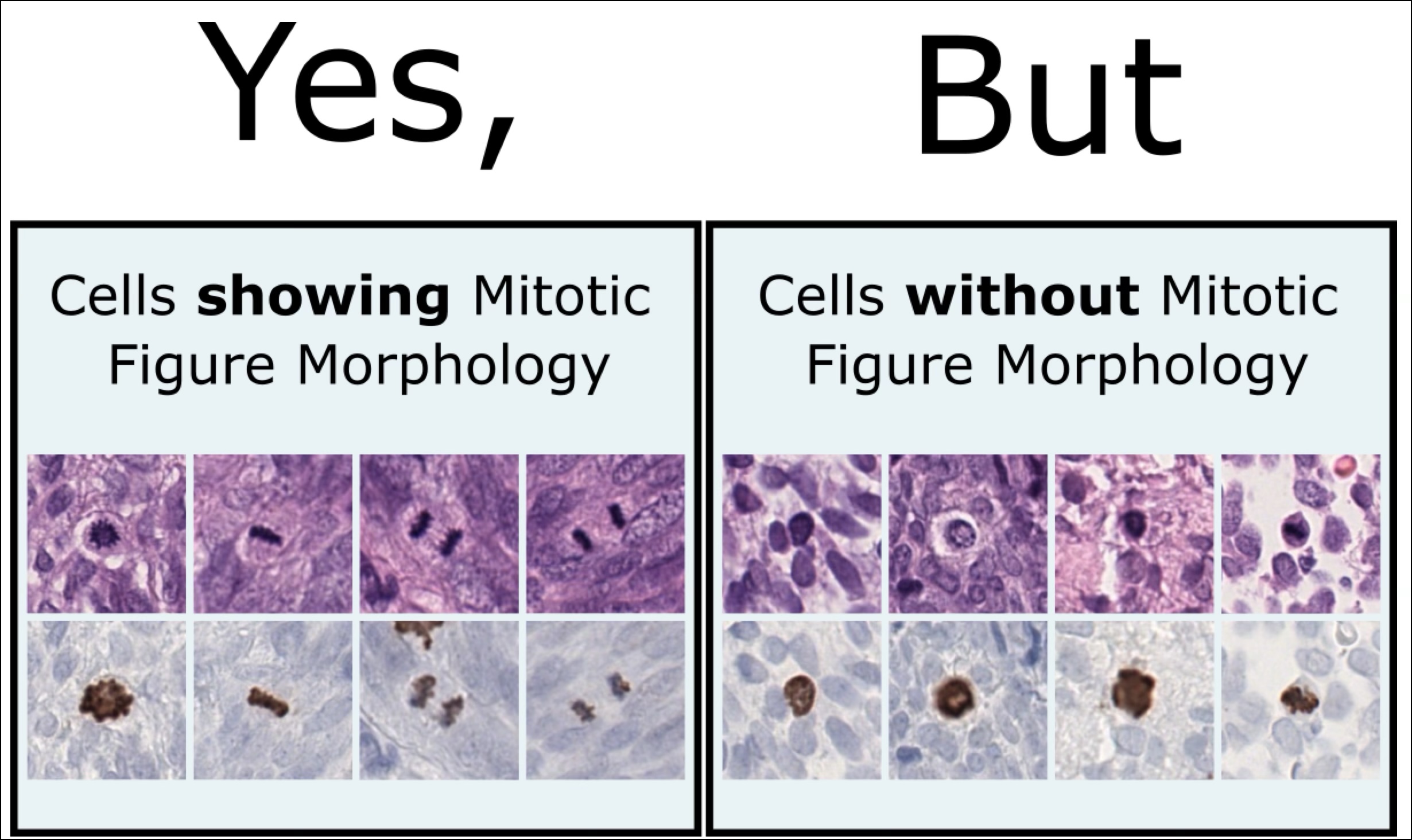
- Information Mismatch in H&E: Despite the increased agreement, the use of PHH3 revealed a key challenge. The PHH3 assistance led to the annotation of many MFs that were difficult or impossible to identify as such in the H&E. This happened even though the pathologists were instructed during the study to annotate only those MFs that were clearly recognizable in the H&E. Some MFs identified with PHH3 lacked clear morphological features in H&E slides, introducing an “interpretation shift” that blurred boundaries for training machine learning models.
AI Models and Label Noise: H&E-based detectors trained on PHH3-assisted labels performed worse than those trained solely on H&E annotations. This result highlights a fundamental mismatch: while PHH3 reveals biological truths, these truths are not always visually apparent in H&E, confusing both pathologists and algorithms. The dual-stain detector on the other hand outperformed its single-stain counterparts. By having access to both stains during training and evaluation, it overcame the information mismatch and achieved higher accuracy across datasets. This shows that the omission of the PHH3 assistance is the cause of the information miss match, which leads to the poorer performance of the single stain models.
Average precision of the single-stain FCOS and dual-stain DI-FCOS models on the test sets, evaluated using five-fold cross-validation on the label sets of the study dataset. Boxes represent the first and third quartiles, lines indicate median values, and plus signs represent mean values across runs. The dual-stain detector significantly outperforms the single-stain detectors on the PHH3-assisted (orange) label sets. The green boxes correspond to a separate experiment conducted on a cleaned label set (ref paper)
Implications: Where Do We Go From Here?
Unfortunately, our results show that even PHH3-assisted annotation does not provide a final solution to the ground truth problem when annotating MFs, as it exposes the annotating pathologist to an hindsight bias. This leads to a dilution of the separation of MFs and MFs look-alikes in the H&E and thus ultimately to more rather than less noise in the data and to poorer model performance.
Nevertheless, PHH3 can be of great use in annotating large data sets. In our work, we have proposed a new annotation workflow in which it is used as a screening tool to identify MF candidates. Because of the high contrast between MFs and background in PHH3 staining, this step could also be done relatively quickly by a single pathologist. However, the final check of each mitosis should be performed by a panel of pathologists in H&E to ensure that there are no information shifts.
References
- (2024): Information mismatch in PHH3-assisted mitosis annotation leads to interpretation shifts in H&E slide analysis. In: Scientific Reports, vol. 14, no. 1, pp. 26273, 2024, ISSN: 2045-2322.


Comments are closed