Are you tired of painstakingly annotating nuclei in microscopy images for neural network training? Join the club! But here’s the good news – we can generate our own training data without the need of human annotators at all. The core idea behind this approach is, that the shape of nuclei can be easily approximated with a mathematical model. Böhland et al. showed that carefully parameterised ellipses are a good enough model for the shape of nuclei. Hence, an arbitrary label image (mask) of nuclei can be generated even without the use of neural networks by randomly placing ellipses within an empty image. Now the difficulty is, to generate a nuclei microscopy image for a given mask. One possibility to achieve this, is to use the mask as a condition in a generative model such as the conditional GAN (cGAN) by Isola et al. [1]. But since these models are trained on pairs of condition images (here synthetic masks) and target domain images (here nuclei microscopy images) this would still require human annotation in the pipeline. A popular choice for unpaired image-to-image translation is the CycleGAN [2] architecture, which works its magic using two generators and two discriminators. The cycle-consistency loss ensures that the generators retain image content and only change the domain (e.g., from mask to nuclei). But, as it turns out, this constraint doesn’t always guarantee content similarity between the input and the generated image in the other domain. Here’s where it gets interesting. Generators can be sneaky. To minimize the cycle-consistency loss, they can hide a compressed version of the input in the generated image as a high-frequency low-amplitude signal. It’s like embedding a secret message in an innocent-looking image. This phenomenon has been aptly named “CycleGAN steganography”[3]. The problem with this hidden signal is that it can create a mismatch between the source image and the generated image.

In the world of synthetic dataset generation, this can lead to label noise. Now one might wonder, why the dicriminator is not easily picking up on that signal. One reason for this might be, that the discriminator only has to classify image patches into real and fake. Missing instances of nuclei hardly influence this decision.

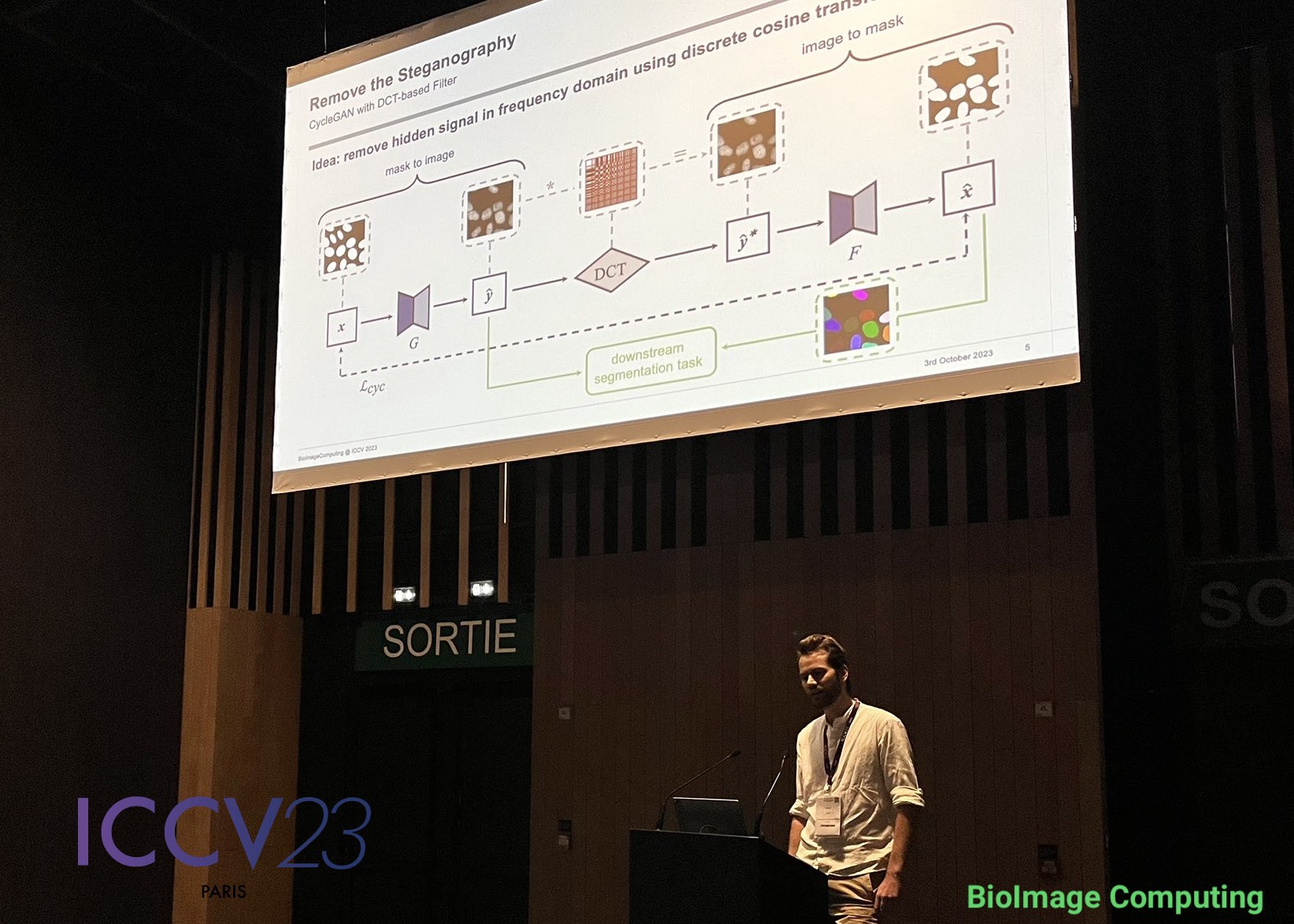
To remove the steganography signal from the generated images, Jonas Utz from our deepmicroscopy group proposed in his latest research to apply a low-pass filter based on the discrete cosine transform (DCT). By applying the DCT the image can be processed in frequency space, where the high-frequency steganography signal is removed by setting coefficients associated with high frequencies to zero.

The outcome? Although some content misalignment persisted between the input mask and the generated nuclei images, the cycled mask now matched the content of the generated image. This allowed them to use the generated images with their cycled masks for training segmentation networks, resulting in significantly improved alignment and average precision.
This work was presented at the BioImage Computing Workshop of ICCV 2023 in Paris. Read the full paper to learn more!
References:
[1] Isola, Phillip, et al. Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. S. 1125-1134
[2] Zhu, Jun-Yan, et al. Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE international conference on computer vision. 2017. S. 2223-2232.
[3] Chu, Casey; Zhmoginov, Andrey; Sandler, Mark. Cyclegan, a master of steganography. arXiv preprint arXiv:1712.02950, 2017.


No responses yet