The advancements in surgical techniques for head and neck cancer have significantly improved patient survival rates. Precision in tumor excision is crucial for successful surgery, and frozen sections are often taken during the procedure. However, the sensitivity of frozen sections is not satisfactory, and extensive sampling poses risks to the patient. Confocal laser endomicroscopy (CLE) has emerged as a non-invasive, real-time imaging modality for Ear-Nose-Throat (ENT) surgeons, offering discriminatory features for tumor delineation. Automatic evaluation of malignancy from CLE images using neural networks has shown promise, especially in regions like the sinunasal cavity, where challenges arise due to limited patient data and high anatomical diversity.
This is an excellent use case for few shot learning (FSL). In a study, to be presented on the German Workshop on Medical Image Computing this year, we trialed some of the most commonly used FSL methods on CLE data. The paper was already made available with Springer.
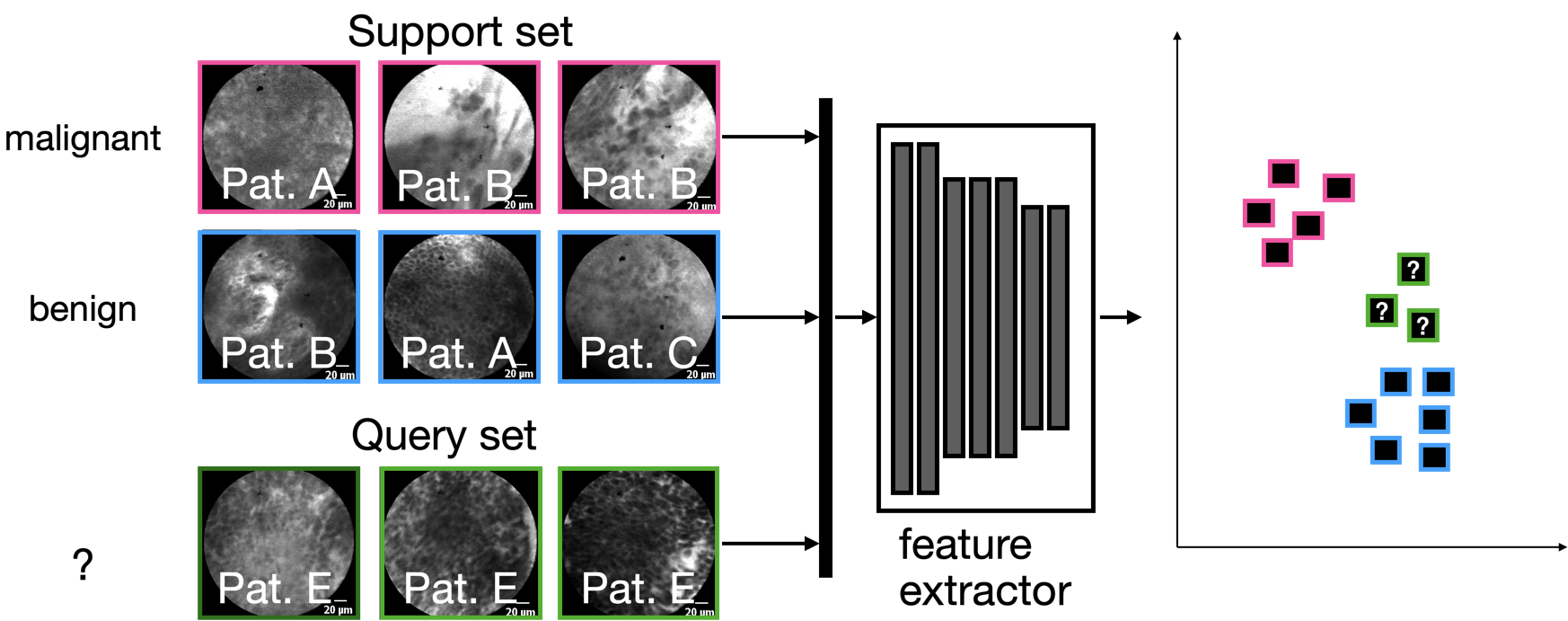
Most FSL methods have in common that they use a feature extractor that was trained specifically to disentangle samples using a support set, which are, however, fixed for the actual dataset. Since only few samples can be utilized for the learning scenario, the only real learning takes place in latent space on the so-called query set, either by using a distance metric, or by training a simple classifier in latent space:
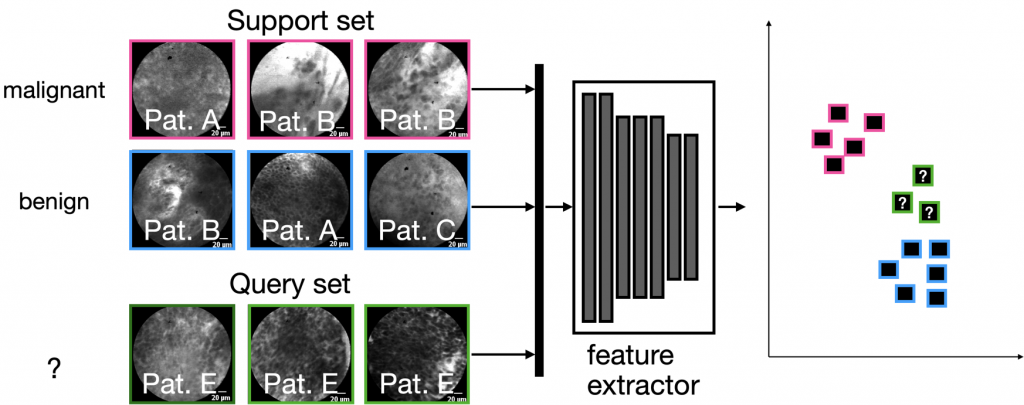
In the particular study, we used CLE images of three different anatomical locations and tumor cases: the oral cavity (squamous cell carcinoma), the vocal folds (squamous cell carcinoma) and the sinunasal cavity (sinunasal tumors).
Our main findings
We found that for the use case of CLE, the inter-patient variability is high – which means that we will need a sufficient number of patients to be able to generalize. This is true for the sinunasal region, where the image diversity is elevated due to the diversity of anatomical structures, but also for the vocal folds, which are by and in itself anatomically more homogeneous.



No responses yet