In the fast-moving world of AI in digital pathology, foundation models (FMs) are making big waves. These powerful models, trained on massive amounts of histopathology data, promise to revolutionize tasks like mitotic figure (MF) classification. But how well do they really perform compared to traditional deep-learning approaches? A recent study by Jonathan Ganz put them to the test, and the results may surprise you.
What’s the Big Deal About Foundation Models?
Foundation models (FM) are trained on vast datasets using self-supervised learning, allowing them to extract high-level features without labeled data. The idea is that these models can then be fine-tuned or used as feature extractors for various medical imaging tasks, potentially reducing the need for large amounts of annotated data. In theory, they should also be more robust to variations in staining techniques, scanners, and other domain shifts in histopathology.
The Experiment: Foundation Models vs. Traditional Training
The researchers compared five publicly available foundation models against conventional deep-learning methods for mitotic figure classification. Specifically, they tested:
- FM-based classifiers using embeddings from models like Virchow, UNI, and Phikon.
- Traditional deep-learning models, including ResNet50 trained end-to-end on labeled MF datasets.
- ImageNet-pretrained models, which use features learned from natural images rather than histopathology data.
They evaluated performance across two large, publicly available MF datasets with varying numbers of labeled examples and assessed how well the models handled domain shifts.
Key Findings: Do Foundation Models Live Up to the Hype?
Surprisingly, the study found that end-to-end trained models outperformed all FM-based classifiers—regardless of how much training data was available. Even with limited data, the traditional models performed better than those relying on embeddings from foundation models.
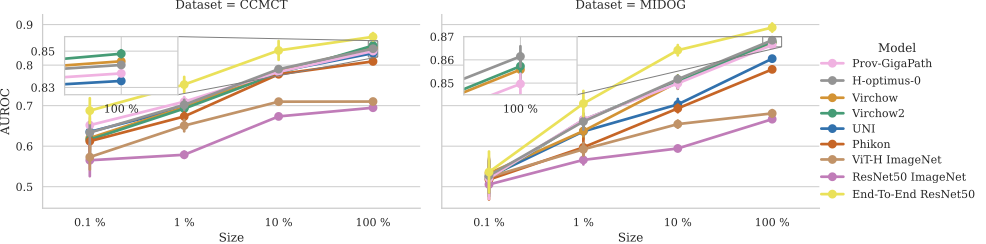
Additionally, one of the key advantages often claimed for FMs—their robustness to domain shifts—didn’t hold up in this study. FM-based classifiers showed no significant improvement in cross-domain performance compared to conventional approaches.
Why Did This Happen?
The findings suggest that while FMs are great at capturing general histopathology features, they may not be optimized for the fine-grained distinctions required in mitotic figure classification. Since MF classification is a highly specialized task, directly training a deep-learning model on the specific dataset still provides the best performance.
Takeaways: Should We Abandon Foundation Models?
Not necessarily! Foundation models still have great potential, especially in settings with very limited labeled data or when tackling a broad range of pathology tasks. However, for highly specific challenges like MF classification, training a dedicated model from scratch still yields the best results.
This study serves as an important reminder that while FMs are a powerful tool, they are not a one-size-fits-all solution. For now, pathologists and AI researchers should continue exploring the best approach for each specific application—sometimes, traditional deep learning still wins the race!
Reference:
Ganz¹, J., Ammeling¹, J., Rosbach¹, E., & Lausser¹, L. (2025, March). Is Self-supervision Enough? Benchmarking Foundation Models Against End-to-End Training for Mitotic Figure Classification. In Bildverarbeitung für die Medizin 2025: Proceedings, German Conference on Medical Image Computing, Regensburg March 09-11, 2025 (p. 63). Springer-Verlag.


Comments are closed