Reducing the annotation effort for microscopy images [MICCAI 2023 paper]
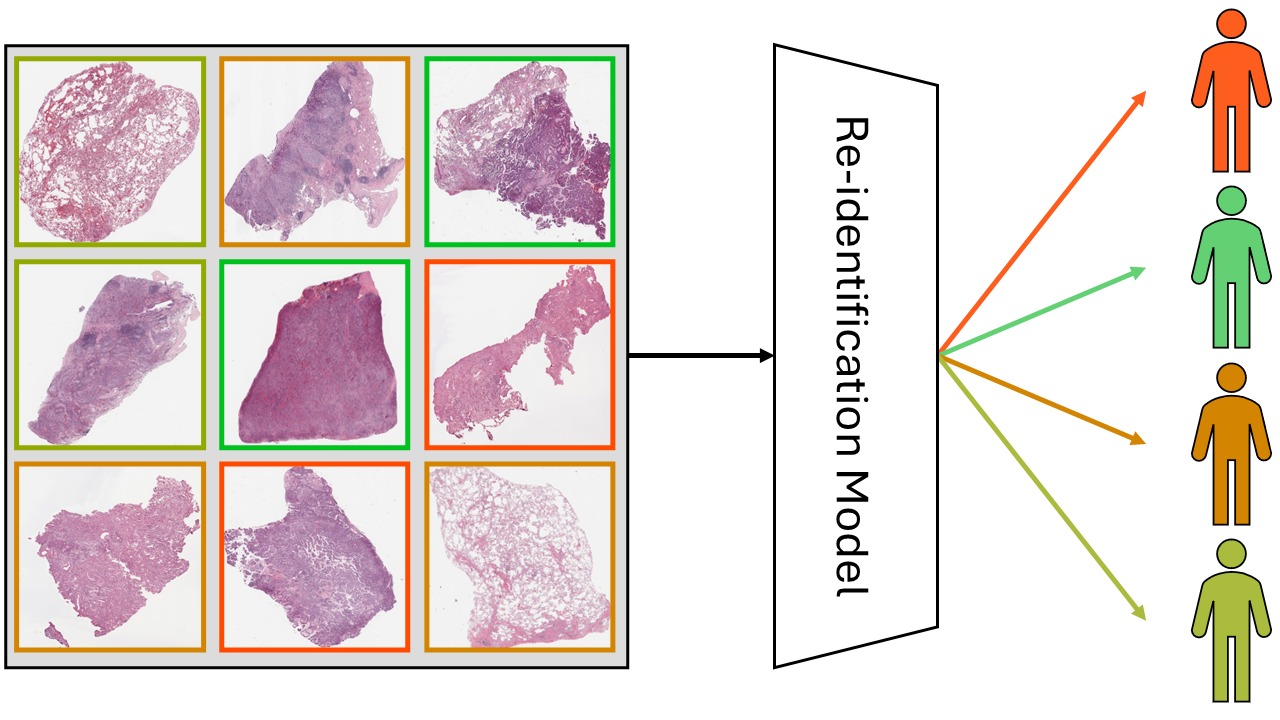
When working with gigantic images – we’re talking 100,000 by 100,000 pixels here – things can get a little tricky. These are known as Whole Slide Images (WSIs), and are often used in medical research. For example, they’re used for studying tissue samples to look for signs of disease. To train a neural network to understand and analyze these WSIs, we need to manually point out and mark various features of interest within the image, a process known as “annotating”.

Image source: “Cats and dogs” (Parkhi, Omkar M., et al.), CAMELYON16 dataset (Litjens, et al.)
Sounds straightforward? Not quite. This process can be time-consuming, especially when you consider the size and complexity of WSIs. Take a widely used breast cancer metastases dataset CAMELYON16 (Litjens, et al.) for instance. In this project, about half the WSIs had important cancerous signs that occupied less than 1% of image. That’s a lot of searching to make sure you don’t miss any small objects of interest. In some other WSIs, on the other hand, the tumor metastases can be so big that drawing their irregular outline can take a substantial amount of time, see the image below.
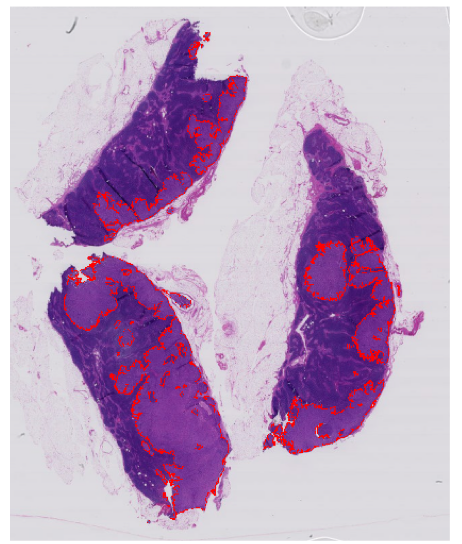
Image source: CAMELYON16 dataset (Litjens et al.)
Luckily, the workload of expert annotators could be greatly reduced if a small number of regions from the WSIs could be found so that the neural network could be properly trained to achieve full annotation performance just with their annotations. The full annotation performance means the performance that the network can achieve by training on the fully annotated dataset. One smart way to find these regions is through “region-based active learning” (Mackowiak, et al.). It selects annotation regions by figuring out “what the network needs” over and over again. For example, one of the most common sampling strategies is called “uncertainty sampling,” and it picks the areas where the model is most uncertain about the class of the sample.
In region-based active learning, we start by randomly picking some regions from each image and annotating them. These annotated areas are then used to train a network, which in turn is used to predict and select other regions in the images that should be annotated next. This iterative cycle of annotating, training, and selecting continues until we reach the desired level of model performance or run out of allocated annotation resources.
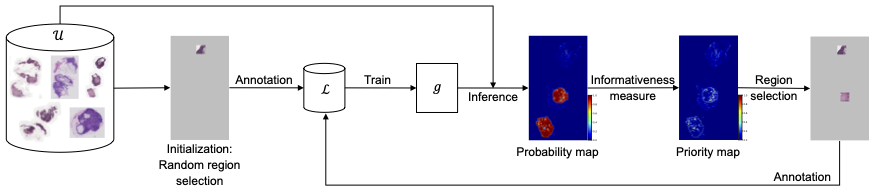
Jingna Qiu from our deepmicroscopy group found in her latest research that the amount of area one chooses to annotate at each active learning cycle, which is typically manually determined based on experience, can have a significant impact on the overall efficiency and cost of the process. If the chosen area is too large or too small, it can lead to inefficiency through extra annotation work or increased computational costs.
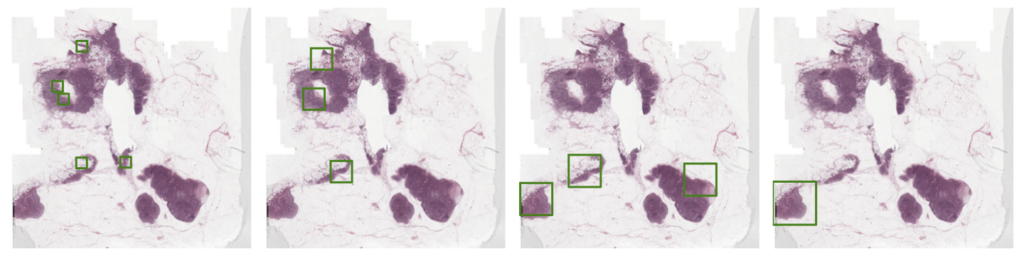
To address this issue, Jingna introduced a new method that adjusts the size of the annotation area dynamically, based on the specifics of the image. This method identifies a potentially informative area and then determines the optimal surrounding area to annotate.
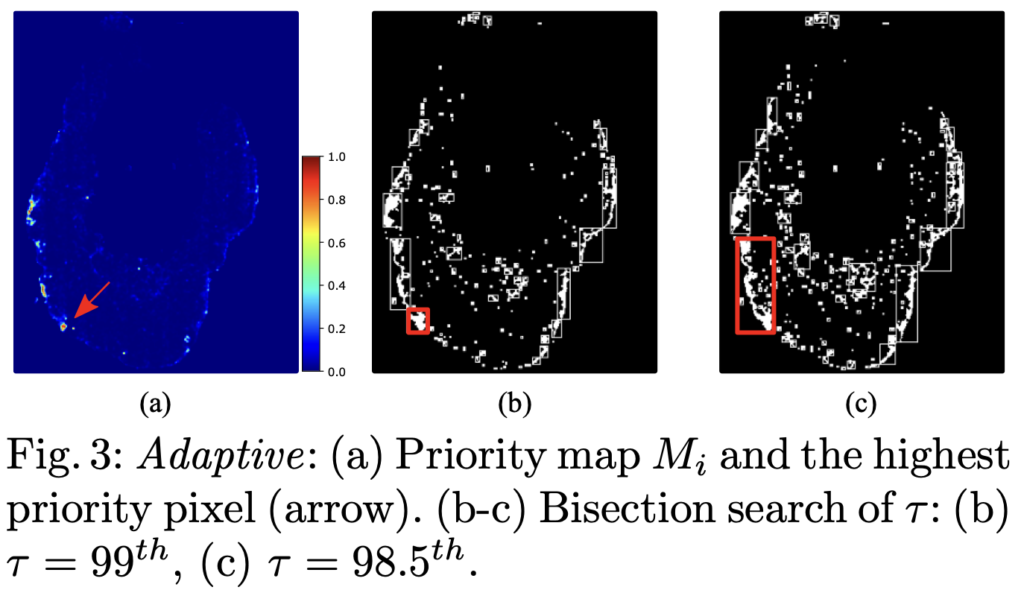
When testing this new method on the well-known CAMELYON16 breast cancer dataset, it proved to be both more efficient and robust than the current standard active learning approaches by achieving the full annotation performance with only 2.6% of the tissue being annotated. That’s a huge time-saver and a potential game-changer for analyzing WSIs.
This work will be presented at this year’s Medical Image Computing and Computer Assisted Intervention (MICCAI 2023) conference in Vancouver. Explore more by reading the paper!
Reference:
[1] Parkhi, Omkar M., Andrea Vedaldi, Andrew Zisserman, and C. V. Jawahar. “Cats and dogs.” In 2012 IEEE conference on computer vision and pattern recognition, pp. 3498-3505. IEEE, 2012.
[2] Litjens, G., Bandi, P., Ehteshami Bejnordi, B., Geessink, O., Balkenhol, M., Bult, P., et al.: 1399 h&e-stained sentinel lymph node sections of breast cancer patients: the camelyon dataset. GigaScience 7(6), giy065 (2018)
[3] Mackowiak, R., Lenz, P., Ghori, O., et al.: Cereals-cost-effective region-based active learning for semantic segmentation. arXiv preprint arXiv:1810.09726 (2018)


No responses yet